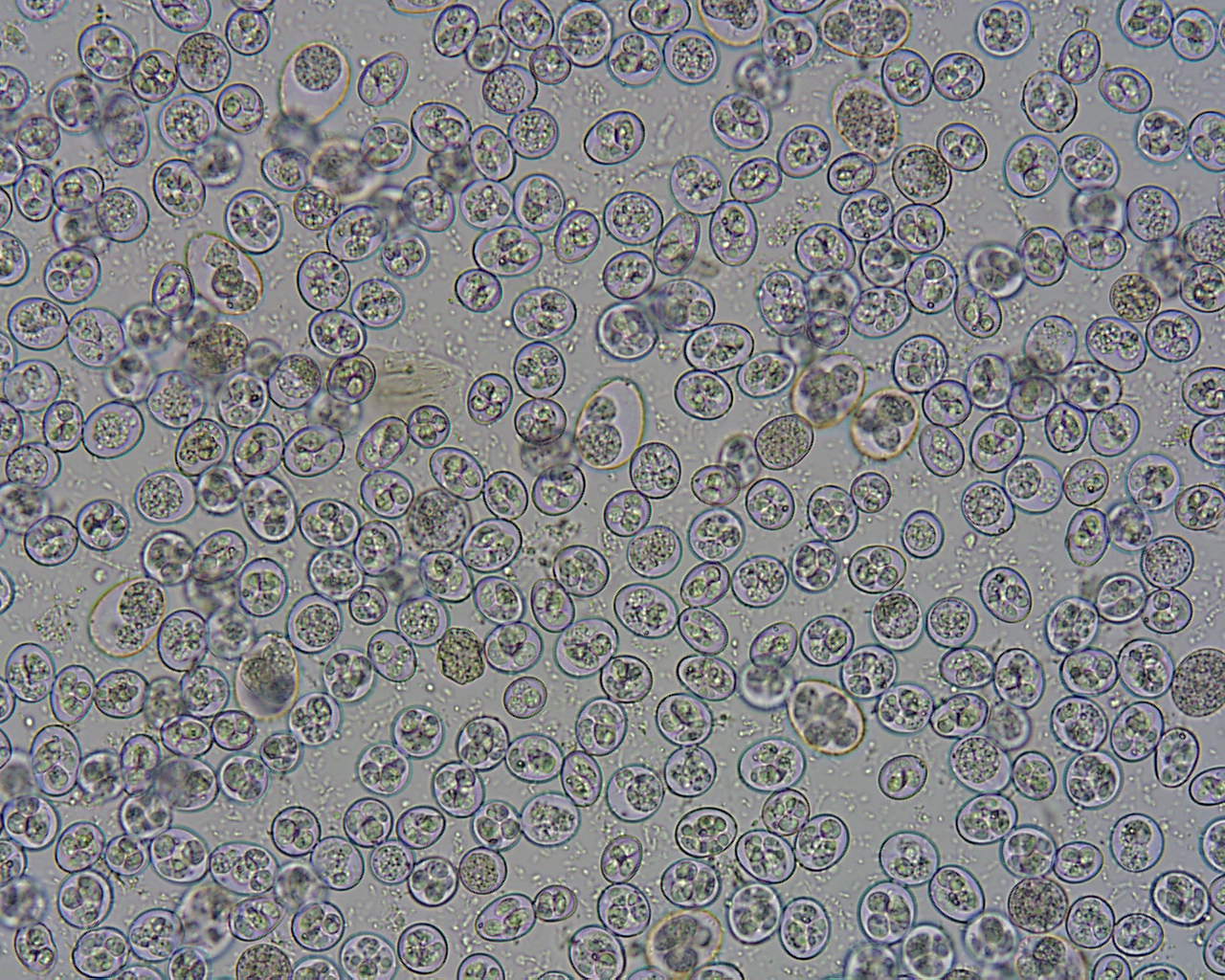
Coccidiosis, caused by parasites of the genus Eimeria, is a worldwide disease of chickens estimated to result in global annual losses of over US$2.4 billion. Ten species of Eimeria have been identified in Australia and species, as well as strains within a species, can vary greatly in their pathogenicity. Recent outbreaks on vaccinated farms have highlighted concerns that the vaccine strains may not be protecting birds sufficiently. Additionally (in Australia), it is not currently possible to distinguish vaccine strains from wild infections.
To assess the nationwide status of coccidiosis, Dr Jess Morgan from the University of Queensland has led a CRC project titled Characterising population structure and diversity of Australian Eimeria. The project, which has recently been completed, sampled species and strains of Eimeria from every Australian state and territory. Different regions of DNA were sequenced to identify variable genetic markers in order to differentiate species and strains of the ten Eimeria species.
Jess explained, “Accurate disease diagnosis is important to the poultry industry where coccidiosis is a costly and virtually ubiquitous problem. The aim of my project was to identify new genetic markers to differentiate among Australian strains of Eimeria. Live Eimeria vaccines have been developed as a means of countering the increasing emergence of drug resistant strains, and although species can be diagnosed using their DNA, there was a need for genetic markers to differentiate among strains of Eimeria”.
During the three year project, five nationwide chicken faecal sampling surveys were conducted. Samples were screened visually and genetically for infective Eimeria oocysts. The high prevalence of uncharacterised Eimeria species in the samples led to the development of a new species-diagnostic genetic assay, targeting a region of mitochondrial DNA. Nationwide samples were screened using the assay comparing backyard and commercial broiler flocks. A subset of strains was passed through laboratory birds to obtain sufficient numbers of oocysts to cryopreserve in a live Eimeria collection.

Jess continued, “DNA regions of the mitochondrial and apicoplast genomes were sequenced from pure Eimeria strains of eight species to identify single nucleotide polymorphisms (SNPs), in order to differentiate among strains. Then, species-specific assays targeting these SNPs were developed so that DNA extracted from mixed species infections could be screened”.
Publicly available nuclear genome sequences for E. tenella and E. maxima were mined for highly repetitive DNA sequences, called microsatellites. During DNA replication, ‘slippage’ of the repeat elements produces differences which can be used to characterise strains. Genetic assays targeting variable microsatellite loci were developed for E. maxima, E. acervulina, E. tenella and E. necatrix.
Australian species and strains were screened using the new strain diagnostic assays in the first nationwide assessment of Eimeria genetic diversity. Strains were genotyped and assigned a genetic barcode. Vaccine strains were also included in the comparison and assigned a barcode.
The project’s outcomes included:
- development of a new assay for species diagnostics that is more comprehensive than any available assay and cheaper than many.
- development of 15 new strain diagnostic assays targeting SNPs in the mitochondrial genome.
- Development of 38 new strain diagnostic assays targeting microsatellites in the nuclear genome of Australian Eimeria species. It is now possible to differentiate vaccine strains from wild strains for four Eimeria species. These assays will reduce the cost of managing outbreaks by providing accurate diagnosis of disease down to the strain level.
- Expansion of the existing library of cryopreserved Eimeria species and strains with the addition of 42 new stabilates containing 123 infections.
The overall conclusion that can be drawn from Jess’ study is that coccidiosis in Australia is widespread, species are ubiquitous, dynamic and genetically diverse, and this diversity does not appear to be geographically structured. Thus national measures to control the spread of coccidiosis do not appear to be necessary.
Jess would like to see future study concentrate on, “Characterising the impact on industry of the three poorly understood operational taxonomic units, measuring the true prevalence of E. tenella in commercial flocks by screening caecal samples, and further testing to optimise vaccine strain diagnostics (with assay development)”.
To obtain a copy of the final report, please contact admin@poultrycrc.com.au.